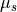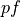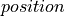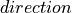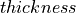Summary¶
Dataset modules contain templates for rendering reference simulation scripts for a number of different sample, source and detector configurations. The reference scripts can be rendered by invoking the appropriate functions or by running the modules. There are several parameters that can be used to control the rendering process when running the modules from a command line:
-oOption can be used to specify a target directory for the rendered scripts and the related datasets. The current working directory is used by default. The scripts will be rendered into
runsubdirectory and the dataset produced by the reference simulation scripts will be saved into thedatadirectory.
-dOption can be used to specify a default OpenCL device that will be used to run the simulations. This value is matched against the vendor and device name (or part of it). The first available OpenCL-enabled GPU device is used by default. Note that the target OpenCL device can be also selected by setting the
CL_DEVICEenvironment variable before running the rendered script.
-iOption can be used to specify the index of the OpenCL device to use. This option is useful when multiple identical devices are installed. Note that the index is zero-based and defaults to 0. Note that the target OpenCL device index can be also selected by setting the
CL_INDEXenvironment variable before running the rendered script.
-vFlag enables verbose output.
-tFlag performs a test run and implicitly enables verbose output. Scripts will be rendered but no files will be written to the target directory. The output will show the full paths of the generated scripts.
The following example will render all the available scripts into the current working directory and produce a verbose output:
python -m xopto.dataset.render.all -v
Details¶
The following sections provide details on different reference datasets and instructions for rendering / generating scripts that will compute these datasets. At the end of each section there is a breakdown of the directory tree and naming conventions that are used to organize the datasets.
MCML comparison datasets¶
Module xopto.dataset.render.mcml_comparison => run/mcml_comparison/
Renders scripts that will produce datasets for comparison with the MCML package.
These datasets are based on
Hg scattering phase function, a
Line source, a
Radial reflectance and
transmittance detector and a FluenceRz
fluence / deposition detector. The refractive index of the surrounding medium
is set to 1.0. The simulation termination radius is set to 1000 mm and each
simulation is run with 100 million photon packets.
The optical properties of the top sample layer are sapled from
![\mu_a = [0.0, 2.5, 5.0]](../../_images/math/0c9106d4f1f6ab37739aa733b351d91118142ce1.png) cm -1,
cm -1,
![\mu_s' = [5.0, 20.0, 35.0]](../../_images/math/58a5c52b70ed9dce542bb251781a8ce168fc8ed7.png) cm -1 and
cm -1 and
![g = [0.1, 0.5, 0.9]](../../_images/math/c396d7426a76ba7ded8685bd54fdbadb757cd7c5.png) .
.
Parameter |
Value |
Description |
|
|---|---|---|---|
normal incidence |
|||
|
|
reflectance and transmittance accumulators |
|
|
|
|
radial |
|
|
|
|
|
‘deposition’ |
weight deposition mode |
run/mcml_comparison/1-layer-100mm.pyA single 100 mm thick sample layer.
A single layer sample of 100 mm thickness¶ Parameter
Value
Description
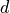
100 mm
thickness
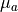
0.0, 2.5 or 5.0 cm -1
absorption coefficient
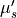
5.0, 20.0 or 35.0 cm -1
reduced scattering coefficient
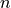
1.337
refractive index
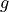
0.1, 0.5 or 0.9
Hg(g)
run/mcml_comparison/2-layer-100um-1mm.pyTwo sample layers, a 0.1 mm top and a 1.0 mm bottom layer. The optical properties are varied only in the top sample layer (the refractive index is fixed to 1.462). The optical properties of the bottom sample layer are fixed to
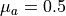 cm -1,
cm -1,
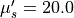 cm -1,
cm -1, 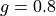 and
and
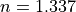 .
.A 2-layer sample¶ Parameter
Value
Description
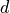
0.1 mm
thickness
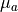
0.0, 2.5 or 5.0 cm -1
absorption coefficient
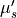
5.0, 20.0 or 35.0 cm -1
reduced scattering coefficient
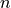
1.462
refractive index
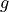
0.1, 0.5 or 0.9
Hg(g)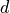
1.0 mm
thickness
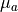
0.5 cm -1
absorption coefficient
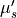
20 cm -1
reduced scattering coefficient
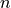
1.337
refractive index
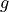
0.8
Hg(g)
Dataset files
Executing the generated scripts will save the simulation results / datasets into compressed numpy data files that will be organized as follows:
data/mcml_comparison/<sample>/line/radial/hg/g-<g>/mua-<mua>-musr-<musr>-invcm.npz
The values of placeholders <> are as follows:
<sample>can take the following values:1-layer-1mmA single layer 1 mm thick medium.1-layer-100mmA single layer 100 mm thick medium.2-layer-100um-1mmA two-layer medium with 0.1 top layer and 1 mm bottom layer
<g>is the anisotropy formatted with two decimal digits and_as the decimal separator, e.g0_10for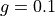 ,
,
0_50for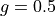 or
or 0_90for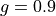 .
.<mua>is the absorption coefficient in units of cm -1 with two decimal digits and_as the decimal separator, e.g2_50for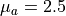 cm -1 .
cm -1 .<musr>is the reduced scattering coefficient in units of cm -1 with two decimal digits and_as a decimal separator, e.g20_00for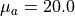 cm -1 .
cm -1 .
Layered media datasets¶
Module xopto.dataset.render.mcml => run/mcml/
Renders scripts that produce datasets for the layered MC kernel. These will include a number of different source, detector and sample scattering phase function configurations. The simulation termination radius is set to 25 mm, except for the spatial frequency domain(SFD) dataset that uses a 150 mm simulation termination radius. The datasets are produced with 100 million photon packets, except for the SFD dataset and all the datasets that use an optical fiber probe with a linear layout of 6 fibers. These datasets are run with 1000 million photon packets. The optical properties of the sample are varied according to the values in the following two tables.
Parameter |
From (cm -1) |
To (cm -1) |
Points |
|---|---|---|---|
|
0.0 |
5.0 |
21 |
|
5.0 |
35.0 |
21 |
Parameter |
Values |
|
|---|---|---|
|
0.1, 0.3, 0.5, 0.7, 0.9 |
|
|
0.1, 0.3, 0.5, 0.7, 0.9 |
|
|
0.0, 0.2, 0.4, 0.6, 0.8, 1.0 |
|
|
0.1, 0.3, 0.5, 0.7, 0.9 |
|
|
-0.5, 0.0, 0.5, 1.0, 1.5 |
|
|
1.337 |
|
|
1.462 (fused silica) |
|
|
0.25, 0.5, 1.0, 2.0, 4.0 μm |
|
|
500 nm |
|
|
1.337 |
|
|
1.603 (polystyrene) |
|
|
0.25, 0.5, 1.0, 2.0, 4.0 μm |
|
|
500 nm |
The refractive index of the sample is set to 1.337.
Reference simulation scripts are rendered for the following basic sources that use a laterally uniform boundary between the sample and the surrounding medium.
Source |
Parameter |
Value |
Reflectance detector |
|---|---|---|---|
|
|||
|
200 μm |
||
|
100 μm |
||
|
200 μm |
||
|
220 μm |
||
|
1.462 |
||
|
0.22 |
The refractive index of the surrounding medium is set to 1.0 except when
using the UniformFiber source,
when the refractive index of the surrounding medium follows the
refractive index of the fiber core 1.462.
The reflectance of basic sources is collected with a radial detector
Radial (Axis (start=0, stop=0.005, n=500))
that extends from 0 to 5 mm with 5oo concentric ring accumulators,
each 5 μm wide.
The acceptance angle is unlimited, except for the
UniformFiber source, for which it is
limited by the NA of the fiber core. The acceptance angle within the
fiber core is computed as 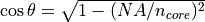 .
.
Reference simulation scripts are also rendered for optical fiber probe
sources that use a surface layout to more accurately describe the interface
between the optical fiber probe tip and the sample.
All the probe sources launch the photon packets with the
UniformFiber source.
Probe |
Parameter |
Value |
Description |
Reflectance detector |
|---|---|---|---|---|
|
200 μm |
six-around-one layout |
||
|
220 μm |
|||
|
1.462 |
|||
|
0.22 |
|||
|
220 μm |
|||
|
6.0 mm |
|||
|
0.6 |
|||
|
400 μm |
six-around-one layout |
||
|
420 μm |
|||
|
1.462 |
|||
|
0.22 |
|||
|
420 μm |
|||
|
6.0 mm |
|||
|
0.6 |
|||
|
200 μm |
linear layout of 6 fibers |
||
|
220 μm |
|||
|
1.462 |
|||
|
0.22 |
|||
|
6 |
|||
|
220 μm |
|||
|
6.0 mm |
|||
|
0.6 |
|||
|
100 μm |
single fiber layout |
||
|
120 μm |
|||
|
1.462 |
|||
|
0.22 |
|||
|
1 |
|||
|
6.0 mm |
|||
|
0.6 |
|||
|
200 μm |
single fiber layout |
||
|
220 μm |
|||
|
1.462 |
|||
|
0.22 |
|||
|
1 |
|||
|
6.0 mm |
|||
|
0.6 |
|||
|
400 μm |
single fiber layout |
||
|
420 μm |
|||
|
1.462 |
|||
|
0.22 |
|||
|
1 |
|||
|
6.0 mm |
|||
|
0.6 |
|||
|
800 μm |
single fiber layout |
||
|
820 μm |
|||
|
1.462 |
|||
|
0.22 |
|||
|
1 |
|||
|
6.0 mm |
|||
|
0.6 |
The reflectance of optical fiber probe sources is collected only through the individual optical fibers of the probe.
SFD source-detector arrangements
The SFD datasets are computed for two source-detector configurations and include raw reflectance and the corresponding frequency-domain reflectance, which is computed for spatial frequencies from 0.00 to 0.80 mm -1 in 0.01 mm -1 steps:
A normally incident
Linesource and a radialRadial(Axis(start=0, stop=0.15, n=4000, logscale=True), cosmin=0.98481) reflectance detector that uses 4000 logarithmically spaced concentric accumulators from 0 to 150 mm. The acceptance angle is limited to 10°. Hankel transform is used to compute the spatial frequency-domain reflectance. Note that this transform produces real values.A normally incident
Linesource and a tilted linear detector with 20° incidence (along the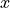 axis).
The accumulators of the detector extend to infinity along the positive
and negative
axis).
The accumulators of the detector extend to infinity along the positive
and negative 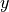 axis and follow a logarithmic spacing along the
positive and negative direction of the
axis and follow a logarithmic spacing along the
positive and negative direction of the 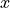 axis
axis
SymmetricX(SymmetricAxis(center=0, range=0.15, n_half=4000, logscale=True), cosmin=0.98480). The described detector uses 8000 (4000 in each direction along the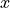 axis) logarithmically spaced accumulators from
axis) logarithmically spaced accumulators from 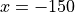 to
to 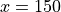 mm. The acceptance angle of the detector
is limited to 10° around the tilted detector axis. Fourier transform is used
to compute the spatial frequency-domain reflectance. Note that this transform
produces complex values with amplitude and phase information.
mm. The acceptance angle of the detector
is limited to 10° around the tilted detector axis. Fourier transform is used
to compute the spatial frequency-domain reflectance. Note that this transform
produces complex values with amplitude and phase information.
Note that the SFD datasets are run with 1000 million photon packets and that the simulation termination radius is set to 150 mm.
Dataset files
Executing the generated scripts will save the simulation results / datasets into compressed numpy data files that will be organized as follows:
data/mcml/<sample>/<source>/<detector>/<pf>/<pf_param_1>/<pf_param_2>/mua-<mua>-musr-<musr>-invcm.npz
The values of placeholders <> are as follows:
<sample>can take the following values:1-layer-semiinfiniteA single samplelayer of infinite thickness.
<source>is the type of the photon packet source / probe used in the datasets:linefor a infinitely narrow line sourceLine.collimated-200umfor aUniformBeamwith a 200 µm beam diameter.gaussian-fwhm-100umfor aGaussianBeamwith 100 µm beam FWHM.fiber-200um-0_22nafor aUniformFiberwith fiber core diameter of 200 µm and 0.22 NA.six-around-one-200um-0_22nafor aSixAroundOneprobe surface layout with a central optical fiber sourceUniformFiber. All optical fibers have a core diameter of 200 µm, cladding diameter of 220 µm, 0.22 NA and are tightly packed.six-around-one-400um-0_22nafor aSixAroundOneprobe surface layout with a central optical fiber sourceUniformFiber. All optical fibers have a core diameter of 400 µm, cladding diameter of 420 µm, 0.22 NA and are tightly packed.six-linear-array-200um-0_22nafor aLinearArrayprobe surface layout with 6 tightly packed optical fibers, a leftmost source fiber (negative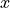 coordinate)
coordinate)
UniformFiber. All optical fibers have a core diameter of 200 µm, cladding diameter of 220 µm, 0.22 NA and are tightly packed.single-fiber-100um-0_22nafor aLinearArrayprobe surface layout with one central optical fiber, a fiber sourceUniformFiber. The optical fiber has a core diameter of 100 µm, cladding diameter of 120 µm, 0.22 NA.single-fiber-200um-0_22nafor aLinearArrayprobe surface layout with one central optical fiber, a fiber sourceUniformFiber. The optical fiber has a core diameter of 200 µm, cladding diameter of 220 µm, 0.22 NA.single-fiber-400um-0_22nafor aLinearArrayprobe surface layout with one central optical fiber, a fiber sourceUniformFiber. The optical fiber has a core diameter of 400 µm, cladding diameter of 420 µm, 0.22 NA.single-fiber-800um-0_22nafor aLinearArrayprobe surface layout with one central optical fiber, a fiber sourceUniformFiber. The optical fiber has a core diameter of 800 µm, cladding diameter of 820 µm, 0.22 NA.
<detector>is the type of detector used by the datasets:radialfor simple sources with laterally uniform source-sample boundary,probefor optical fiber probes with surface layout.
<pf>is the type of scattering phase function used in the datasets:pf_param_1: is the first parameter of the scattering phase function formatted with two decimal digits and using_as the decimal separator:pf_param_2: is the second parameter of the scattering phase function formatted with two decimal digits and using_as the decimal separator. An exception to this rule is the wavelength parameter of theMiescattering phase function that is converted to nm and formatted as an integer. This placeholder is not used with theHgscattering phase function.<mua>is the absorption coefficient in units of cm -1 with two decimal digits and_as the decimal separator, e.g2_50for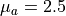 cm -1.
cm -1.<musr>is the reduced scattering coefficient in units of cm -1 with two decimal digits and_as a decimal separator, e.g20_00for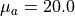 cm -1.
cm -1.
Voxelized media datasets¶
Module xopto.dataset.render.mcvox => run/mcvox/
Renders a dataset scripts for computing fluence / deposition
datasets with the MC kernel for voxelized media. A two-layer skin model
with an embedded blood vessel is used. The depth/position of the blood vessel
along the  axis is varied from 0.2 to 0.8 mm in steps of
0.025 mm.
The refractive index of the surrounding medium is set to 1.337.
The simulations are run with 1000 million photon packets.
axis is varied from 0.2 to 0.8 mm in steps of
0.025 mm.
The refractive index of the surrounding medium is set to 1.337.
The simulations are run with 1000 million photon packets.
Parameter |
Value |
Description |
|
|---|---|---|---|
normally incident |
|||
|
|
16.5724 cm -1 |
epidermis |
|
375.9398 cm -1 |
||
|
1.337 |
||
|
|
||
|
|
45.85 cm -1 |
dermis |
|
356.5406 cm -1 |
||
|
1.337 |
||
|
|
||
|
|
230.5427 cm -1 |
blood vessel |
|
93.985 cm -1 |
||
|
1.337 |
||
|
|
||
|
|
|
sample and deposition voxelization |
|
|
||
|
|
||
|
|
reflectance and transmittance detectors |
|
|
|
||
Blood vessel |
|
200.0 μm |
in dermis |
|
(0, 0, 0.0002-0.0008) |
(x, y, z) |
|
|
(0, 1, 0) |
(x, y, z) |
|
Epidermis |
|
100 μm |
Dataset files
Executing the generated scripts will save the simulation results / datasets into compressed numpy data files that will be organized as follows:
data/mcvox/fluence/2-layer-skin-<diameter>um-vessel-<depth>um-depth-deposition.npz
The values of placeholders <> are as follows:
<diameter>is the diameter of the blood vessel in units of μm, formatted as an integer value, e.g200for a 200 μm blood vessel.<depth>is the coordinate (depth) of the blood vessel in units
of μm, formatted as an integer value, e.g
coordinate (depth) of the blood vessel in units
of μm, formatted as an integer value, e.g 500for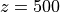 μm.
μm.
Sampling volume datasets¶
Module xopto.dataset.render.sv => run/sv/
Renders a reference dataset script for computing sampling-volume datasets. The sampling volume dataset is computed for a semi-infinite homogeneous medium for an optical fiber probe with two optical fibers placed at a distance of 0.5 mm. The refractive index of the surrounding medium is set to 1.0. Simulations are run in batches until 1,000,000 photon packet traces that reach the detector fiber are collected and converted to sampling volume information. The trace capacity is limited to 1000 events. The simulation termination radius is set to 25 mm.
Parameter |
Value |
Description |
|
|---|---|---|---|
|
200 μm |
linear layout of 2 fibers |
|
|
220 μm |
||
|
1.462 |
||
|
0.22 |
||
|
500 μm |
||
|
2 |
||
|
6.0 mm |
||
|
0.6 |
||
|
1000 |
packet trace configuration |
|
|
|
sampling volume voxelization |
|
|
|
||
|
|
||
|
2.0 cm -1 |
sample layer |
|
|
500.0 cm -1 |
||
|
1.337 |
||
|
|
Dataset files
Executing the generated scripts will save the simulation results / datasets into compressed numpy data files that will be organized as follows:
data/mcml/1-layer-semiinfinite/sv/reflectance/fiber-200um-0_22na/sds-<sds>um/hg/g-<g>/um-mua-<mua>-musr-<musr>-invcm.npz
The values of placeholders <> are as follows:
<sds>is the distance between the centers of the source and detector fibers in units of μm, formatted as an integer value, e.g500for a 500 μm distance.<g>is the anisotropy formatted with two decimal digits and_as the decimal separator, e.g0_15for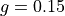 .
.<mua>is the absorption coefficient in units of cm -1 with two decimal digits and_as the decimal separator, e.g2_50for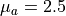 cm -1 .
cm -1 .<musr>is the reduced scattering coefficient in units of cm -1 with two decimal digits and_as a decimal separator, e.g20_00for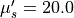 cm -1.
cm -1.
All available datasets¶
xopto.dataset.render.allRenders all the available dataset scripts.
 axis
axis
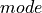
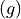
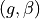
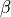
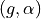
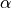
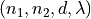
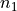
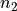
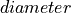
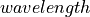
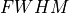
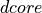
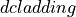
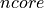
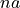
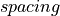
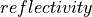
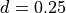 µm.
µm.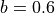 .
.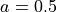 .
.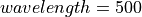 nm.
nm.