Voxelized Monte Carlo simulations¶
In this example we simulate the deposited energy in a voxelized two-layered skin model comprising epidermis and dermis with an embedded blood vessel.
Importing the required modules and submodules¶
From xopto.mcvox we import the module mc, which enables an interface to Monte Carlo voxelized simulations, definitions of voxel grids, fluence accumulators, materials and so on, from xopto.mcvox.mcutil we import module xopto.mcvox.mcutil.axis, which allows axes definitions for voxels and fluence accumulators and finally, we import the module xopto.cl.clinfo from xopto.cl for computational device definition. Additionally, we require modules numpy and matplotlib.pyplot for mathematical functions and visualization.
from xopto.mcvox import mc
from xopto.mcvox.mcutil import axis
from xopto.cl import clinfo
import numpy as np
import matplotlib.pyplot as pp
Computational device¶
In this section we select the desired OpenCL computational device (see also OpenCL devices).
cl_device = clinfo.gpu(platform='nvidia')
Note
In this example we have selected the first computational device listed under the Nvidia platform. The string should be changed according to the installed hardware devices.
Voxel grid and tissue structure¶
For the definition of the voxel grid, we firstly define the number of bins in each axis direction nx, ny and nz. We chose odd number of bins nx and ny in order to have the center of the central voxel at 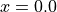 and
and 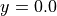 . Using the
. Using the Axis constructor, we define the axes in x, y and z direction. These are defined in a way that there is an equal number of voxels on each side of the origin in the x and y direction and a single central voxel. Note that the leftmost and rightmost edges are provided as keyword arguments start and stop and that the rightmost edge is included in the provided interval. The keyword parameter n provides the number of bins. The voxel object is then defined using the Voxels contructor by passing the axes.
# number of bins and size
nx = 201
ny = 201
nz = 200
binsize = 5e-6
# define each axis
xaxis = axis.Axis(
start=-nx/2 * binsize,
stop=nx/2 * binsize,
n=nx
)
yaxis = axis.Axis(
start=-ny/2 * binsize,
stop=ny/2 * binsize,
n=ny
)
zaxis = axis.Axis(
start=0.0,
stop=nz * binsize,
n=nz
)
# define voxels
voxels = mc.mcgeometry.Voxels(
xaxis=xaxis,
yaxis=yaxis,
zaxis=zaxis
)
Once the voxels are defined, we can set the structure of the tissue. This can be easily done by logical indexing that is readily available within the numpy library. Firstly, we can obtain x, y, z coordinate matrices or a mesh by calling a meshgrid method. These can then be used to impose geometrical logical conditions under which we can specify optical properties only to selected voxels. In this example, the tissue will resemble human skin comprising epidermis, dermis and a blood vessel. The blood vessel can be desribed by a cylinder of radius r_vessels equal to 100 μm placed along the y axis at x0 and z0. The depth z0 is at half of the total thickness of the voxelized medium in z direction.
Subsequently, we define logical matrices of the same shape and size as the voxelized medium mesh. The matrix of logical indices logical_epi corresponds to the epidermis and has True values only for z coordinates lower than 60 μm, logical_der corresponds to the dermis having True values beyond 60 μm. Finally, the matrix of logical indices logical_vessel contains logical True values within the cylinder representing the blood vessel. These logical matrices will be utilized to assign the optical properties later in this example.
# get mesh
z, y, x = voxels.meshgrid()
# set logical indices for material assignment
r_vessels = 100e-6
x0 = 0.0
z0 = nz/2 * binsize
logical_epi = z <= 60e-6
logical_der = z > 60e-6
logical_vessel = (x-x0)**2 + (z-z0)**2 <= r_vessels**2
Materials¶
Each material that will be utilized within the structured tissue can be constructed using the Material class, which accepts refractive index n, absorption coefficient mua in 1/m, scattering coefficient mua in 1/m and scattering phase function. Various scattering phase functions that are available in the package xopto.mcbase.mcpf can be chosen. In this case we select Henyey-Greenstein phase function for all materials. We define optical properties for surrounding medium material_sm, epidermis material_epi, dermis material_der and blood material_bl. These values were obtained from the Steven Jacques’ example for mcxyz under the section How to use mcxyz.c. Finally, it should be noted that the defined materials have to be packed by using the :py:class`~xopto.mcvox.mcmaterial.Materials` constructor, which accepts a lsit of Material objects and is later in this example passed to the Monte Carlo simulator constructor Mc. Note that the order of the Material objects is important, since voxels will be assigned indices that correspond to the order of these materials in the list. Initially, all voxels are assigned an index 0, which corresponds to the first material or in our case the surrounding medium material_sm. The proper material assignment to each voxel will be done later in this example.
# surrounding medium
material_sm = mc.mcmaterial.Material(
n=1.33,
mua=0.0001e2,
mus=1.0e2,
pf=mc.mcpf.Hg(1.0)
)
# epidermis
material_epi = mc.mcmaterial.Material(
n=1.33,
mua=16.5724e2,
mus=375.9398e2,
pf=mc.mcpf.Hg(0.9)
)
# dermis
material_der = mc.mcmaterial.Material(
n=1.33,
mua=0.4585e2,
mus=356.5406e2,
pf=mc.mcpf.Hg(0.9)
)
# blood
material_bl = mc.mcmaterial.Material(
n=1.33,
mua=230.5427e2,
mus=93.9850e2,
pf=mc.mcpf.Hg(0.9)
)
materials = mc.mcmaterial.Materials([
material_sm,
material_epi,
material_der,
material_bl
])
Source¶
In this example we define a pencil beam source, which can be initialized using the Line class. Two keyword parameters can be passed position and direction. In this case we keep the default values which describe the source position at the origin and perpendicular to the surface of the medium.
source = mc.mcsource.Line()
Fluence object¶
To accumulate either fluence or deposited energy, we have to define an instance of Fluence. The Fluence class accepts x, y and z axes and a mode parameter, which can be assigned a string fluence or deposition, depending on the desired simulated quantity stored in the fluence object. In case the mode is fluence, the accumulated quantity is fluence with units of 1/m:superscript:2, under the mode deposition, the accumulated quantity is the energy deposition in a voxel with units 1/m:superscript:3 and normalized against the initial number of launched photon packets. To obtain physically representable energy deposition units, the accumulated values should be multiplied by the power of the source in Watts.
fluence = mc.mcfluence.Fluence(
xaxis=xaxis,
yaxis=yaxis,
zaxis=zaxis,
mode='deposition'
)
The Monte Carlo simulator object and material assignment¶
In this section we define the Mc simulator object, which accepts various parameters and subsequently assign materials to voxels. Mandatory parameters to the Mc simulator object constructor are voxels, materials and source, which we have already defined in the previous sections. We also pass the fluence object fluence to accumulate the deposited energy and also the cl_device object to specify the computational device on which the simulations are going to be run.
Once the Mc simulator object is defined, we can assign the materials to voxels. We use the previously defined logical matrices corresponding to the different tissue structures to access the voxels and assign indices that correspond to the order of materials packed in the materials object.
mc_obj = mc.Mc(
voxels=voxels,
materials=materials,
fluence=fluence,
source=source,
cl_devices=cl_device,
)
# assign the materials to voxels
mc_obj.voxels.material[logical_epi] = 1
mc_obj.voxels.material[logical_der] = 2
mc_obj.voxels.material[logical_vessel] = 3
Running the Monte Carlo simulation¶
To run the Monte Carlo simulations, we set the number of photons to 10:superscript:8 and call the method run(), which returns three objects. The fluence is returned as the second object, which is thus achieved by [1].
nphotons = 1e8
deposit = mc_obj.run(nphotons, verbose=True)[1]
Visualize the results¶
Finally, we visualize the obtained deposited energy, which can be accessed through the attribute data in the returned Fluence object deposit. The visualization is done in the logarithmic scale. Note that visualization can also be done in a straightforward manner by calling the method plot() and specifying the axis along which slices through the 3D medium are performed.
# plot the deposited energy in 1/m^3.
pp.figure()
pp.title('Deposited energy')
extent = [1e3*(xaxis.centers[0] - binsize/2), 1e3*(xaxis.centers[-1] - binsize/2),
1e3*(zaxis.centers[-1] + binsize/2), 1e3*(zaxis.centers[0] - binsize/2)
]
pp.imshow(np.log10(deposit.data[:,100,:]),
extent=extent,
origin='upper'
)
pp.xlabel('x (mm)')
pp.ylabel('z (mm)')
cbar = pp.colorbar()
cbar.set_label('Log10(A)')
pp.show()
# use the slicer view
deposit.plot(axis='y')
The complete example¶
# -*- coding: utf-8 -*-
################################ Begin license #################################
# Copyright (C) Laboratory of Imaging technologies,
# Faculty of Electrical Engineering,
# University of Ljubljana.
#
# This file is part of PyXOpto.
#
# PyXOpto is free software: you can redistribute it and/or modify
# it under the terms of the GNU General Public License as published by
# the Free Software Foundation, either version 3 of the License, or
# (at your option) any later version.
#
# PyXOpto is distributed in the hope that it will be useful,
# but WITHOUT ANY WARRANTY; without even the implied warranty of
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
# GNU General Public License for more details.
#
# You should have received a copy of the GNU General Public License
# along with PyXOpto. If not, see <https://www.gnu.org/licenses/>.
################################# End license ##################################
# This example shows how to simulate deposited energy using the
# voxelized Monte Carlo method for a simple case of a two-layered
# skin model with an embedded blood vessel.
from xopto.mcvox import mc
from xopto.cl import clinfo
import numpy as np
import matplotlib.pyplot as pp
# DEFINE A COMPUTATIONAL DEVICE
# select the first device from the provided platform
cl_device = clinfo.gpu(platform='nvidia')
# DEFINE VOXELS AND TISSUE STRUCTURE
# number of bins and size
nx = 201
ny = 201
nz = 200
binsize = 5e-6
# define each axis
xaxis = mc.mcgeometry.Axis(
start=-nx/2 * binsize,
stop=nx/2 * binsize,
n=nx
)
yaxis = mc.mcgeometry.Axis(
start=-ny/2 * binsize,
stop=ny/2 * binsize,
n=ny
)
zaxis = mc.mcgeometry.Axis(
start=0.0,
stop=nz * binsize,
n=nz
)
# define voxels
voxels = mc.mcgeometry.Voxels(
xaxis=xaxis,
yaxis=yaxis,
zaxis=zaxis
)
# get mesh
z, y, x = voxels.meshgrid()
# set logical indices for material assignment
r_vessels = 100e-6
x0 = 0.0
z0 = nz/2 * binsize
logical_epi = z <= 60e-6
logical_der = z > 60e-6
logical_vessel = (x-x0)**2 + (z-z0)**2 <= r_vessels**2
# DEFINE MATERIALS
# surrounding medium
material_sm = mc.mcmaterial.Material(
n=1.33,
mua=0.0001e2,
mus=1.0e2,
pf=mc.mcpf.Hg(1.0)
)
# epidermis
material_epi = mc.mcmaterial.Material(
n=1.33,
mua=16.5724e2,
mus=375.9398e2,
pf=mc.mcpf.Hg(0.9)
)
# dermis
material_der = mc.mcmaterial.Material(
n=1.33,
mua=0.4585e2,
mus=356.5406e2,
pf=mc.mcpf.Hg(0.9)
)
# blood
material_bl = mc.mcmaterial.Material(
n=1.33,
mua=230.5427e2,
mus=93.9850e2,
pf=mc.mcpf.Hg(0.9)
)
materials = mc.mcmaterial.Materials([
material_sm,
material_epi,
material_der,
material_bl
])
# DEFINE PENCIL BEAM SOURCE
source = mc.mcsource.Line()
# DEFINE FLUENCE OBJECT
fluence = mc.mcfluence.Fluence(
xaxis=xaxis,
yaxis=yaxis,
zaxis=zaxis,
mode='deposition'
)
# DEFINE MC OBJECT FOR MONTE CARLO SIMULATIONS AND ASSIGN MATERIALS
mc_obj = mc.Mc(
voxels=voxels,
materials=materials,
fluence=fluence,
source=source,
cl_devices=cl_device,
)
# assign the materials to voxels
mc_obj.voxels.material[logical_epi] = 1
mc_obj.voxels.material[logical_der] = 2
mc_obj.voxels.material[logical_vessel] = 3
# RUN THE MONTE CARLO SIMULATIONS
nphotons = 1e8
deposit = mc_obj.run(nphotons, verbose=True)[1]
# VISUALIZE THE RESULTS
# plot the deposited energy in 1/m^3.
pp.figure()
pp.title('Deposited energy')
extent = [1e3*(xaxis.centers[0] - binsize/2), 1e3*(xaxis.centers[-1] - binsize/2),
1e3*(zaxis.centers[-1] + binsize/2), 1e3*(zaxis.centers[0] - binsize/2)
]
pp.imshow(np.log10(deposit.data[:,100,:]),
extent=extent,
origin='upper'
)
pp.xlabel('x (mm)')
pp.ylabel('z (mm)')
cbar = pp.colorbar()
cbar.set_label('Log10(A)')
pp.show()
# use the slicer view
deposit.plot(axis='y')
You can run this example from the root directory of the PyXOpto package as:
python examples/mcvox/voxelized_vessel.py